You can find Vpython at http://vpython.org/contents/download_windows.html
NB: later we will use a much more powerfull math library such Gnuplot,Python(x,y) but now we will stick to the Vpython.
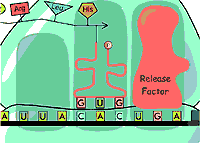
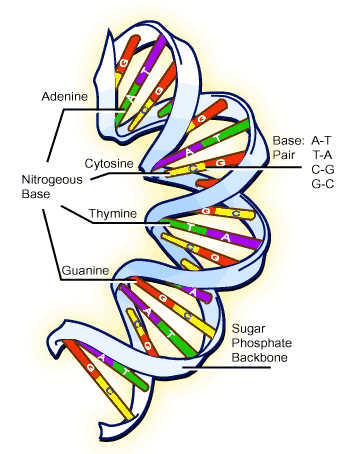
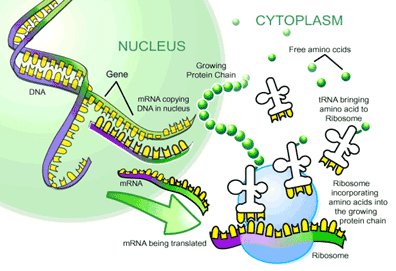
Since we have already enumearted our aminoacid (See the Translation Step ),we had just to enumerate the nucleotids in the DNA fragment so:
- A_nb=0
- C_nb=0
- G_nb=0
- T_nb=0
- for i in range(len(ADN_5)):
- if ADN_5[i]=='A':
- A_nb=A_nb + 1
- if ADN_5[i]=='C':
- C_nb=C_nb + 1
- if ADN_5[i]=='G':
- G_nb=G_nb + 1
- if ADN_5[i]=='T':
- T_nb=T_nb + 1
- print "A_nb =" , A_nb
- print "C_nb =" , C_nb
- print "G_nb =" , G_nb
- print "T_nb =" , T_nb
We have nucleotides and aminoacid frequency in this DNA so let do a simple colored graph from this data
First we need the Vpython library that must be called at the beginning of our code source by this syntax:
- from visual.graph import *
- graph1 = gdisplay(foreground=color.black, background=color.white)
each nuclotid number (A_nb,C_nb,G_nb,T_nb) will be represented in a vertical bar (gvbars ) with different color (color=color.XXXX) and will be displayed at a specific positon of the graph.
the 4 nuclotode are represented in graph using:
- gvbars(delta=0.05, color=color.blue).plot(pos=(0.2,A_nb))
- gvbars(delta=0.05, color=color.red).plot(pos=(.4,C_nb))
- gvbars(delta=0.05, color=color.green).plot(pos=(.6,G_nb))
- gvbars(delta=0.05, color=color.yellow).plot(pos=(.8,T_nb))
- graph2 = gdisplay(foreground=color.black, background=color.white)
- gvbars(delta=0.05, color =(.0,.0,.3)).plot(pos=(.1,Phe_nb))
- gvbars(delta=0.05, color =(.0,.0,.6)).plot(pos=(.2,Leu_nb))
- gvbars(delta=0.05, color =(.0,.0,.9)).plot(pos=(.3,Iso_nb))
- gvbars(delta=0.05, color =(.0,1,.0)).plot(pos=(.4,Met_nb))
- gvbars(delta=0.05, color =(.0,1,.3)).plot(pos=(.5,Val_nb))
- gvbars(delta=0.05, color =(.0,1,.6)).plot(pos=(.6,Ser_nb))
- gvbars(delta=0.05, color =(.0,1,.9)).plot(pos=(.7,Pro_nb))
- gvbars(delta=0.05, color =(1,.0,.0)).plot(pos=(.8,Thr_nb))
- gvbars(delta=0.05, color =(1,.0,.3)).plot(pos=(.9,Ala_nb))
- gvbars(delta=0.05, color =(1,.0,.6)).plot(pos=(1,Tyr_nb))
- gvbars(delta=0.05, color =(1,.0,.9)).plot(pos=(1.1,His_nb))
- gvbars(delta=0.05, color =(1,.3,.0)).plot(pos=(1.2,Gln_nb))
- gvbars(delta=0.05, color =(1,.6,.0)).plot(pos=(1.3,Asn_nb))
- gvbars(delta=0.05, color =(1,.9,.0)).plot(pos=(1.4,Lys_nb))
- gvbars(delta=0.05, color =(1,.3,.3)).plot(pos=(1.5,Asp_nb))
- gvbars(delta=0.05, color =(1,.3,.6)).plot(pos=(1.6,Glu_nb))
- gvbars(delta=0.05, color =(1,.3,.9)).plot(pos=(1.7,Cys_nb))
- gvbars(delta=0.05, color =(1,.6,.0)).plot(pos=(1.8,Trp_nb))
- gvbars(delta=0.05, color =(1,.6,.3)).plot(pos=(1.9,Arg_nb))
- gvbars(delta=0.05, color =(1,.6,.9)).plot(pos=(2,Gly_nb))
- gvbars(delta=0.05, color =(1,.9,.0)).plot(pos=(2.1,STOP_nb))
I hope you enjoyed this little project the whole code source is availble right here